쿡북에 있는 것 중 일부 생략했다. (BLAT이랑 8.3~8.5부분)
BLAST는 Basic local alignment search tool의 준말로, 보통 블래스트라고 한다. 지나가는 2~30대를 붙잡고 크흐~대 기억이~ 하면 지~난 사랑이~ 가 나오듯, 지나가던 생물학도를 붙잡고 블래스트? 하면 NCBI! 하는 정도로 국민 툴이고 전공수업을 듣다 보면 한 번은 쓰고 넘어가는 툴이기도 하다. 애초에 전공이 생물학인지를 먼저 물어봐야 하는 거 아니냐
이 글에 있는 게 어렵다… 그러면 조용히 구글을 열고 BLAST 검색해서 웹사이트 들어가자. 거기서 할 수 있다.
NCBIWWW
www가 우리말로 ㅋㅋㅋ이긴 한데 그건 일본에서 통하는 말이고... 보통은 월드 와이드 웹이다. BLAST를 따로 깔지 않고도 돌리려면 저게 있어야 한다. 물론 BLAST 깔려있는 서버가 있으면 저거 쓰지 말고 걍 로컬 돌리자. 저거 생각보다 느리다. 어느정도로 느리냐면 돌려놓고 강의실 있는 건물에 있는 카페에서 커피 한 잔 마시고 오면 끝난다. 돌려놓고 설거지 한번 하면 된다
from Bio.Blast import NCBIWWW아무튼 오늘은 이 친구를 필두로 뭘 또 많이 데려올 예정이다.
인터넷으로 BLAST 돌리기-기본편
참고로 로컬로 돌리는 건 생략. 놋북에 그게 안 깔려있다.
from Bio.Blast import NCBIWWW
result_handle = NCBIWWW.qblast("blastn", "nt", "8332116")
print(result_handle)기본 코드는 이거다. 이걸 실행하면 더럽게 오래 걸리면서...
<_io.StringIO object at 0x7f2f672ff280>이러시는 이유가 있으실 것 아니예요.
쟤가 왜 저러는지는 이따 저장할 때 설명하기로 하고, 저 세 개가 뭔지에 대해 알아보자. Biopython에서 BLAST를 돌릴 때는 qblast()를 쓰는데, 거기서 필요한 3대 요소가 뭘로 돌릴래/데이터베이스 어디서 찾을래/GI 번호이다. GI 번호는

이런 식으로 Genbank에서 뭔가를 찾으면 나오는 번호. 즉 저 코드는 GI 번호가 8332116인 유전자의 시퀀스로 blastn을 돌리라는 얘기다. (DB 저거 약어같은데 나도 뭔지 모름) 혹시 여기에 대해 더 궁금한 점이 있다면
한번 읽어보자. NCBI에서 제공하는 가이드이다. (물론 영어다)
인터넷으로 BLAST 돌리기-FASTA 파일로 돌리기
Genbank도 돌릴 수는 있는데 뭐가 문젠지 결과를 조졌다... 결과 파일에 암것도 없음.
from Bio.Blast import NCBIWWW
fasta_string = open("/home/koreanraichu/CaMV.fasta").read()
result_handle = NCBIWWW.qblast("blastn", "nt", fasta_string)
print(result_handle)from Bio import SeqIO
from Bio.Blast import NCBIWWW
record = SeqIO.read("/home/koreanraichu/CaMV.fasta", format="fasta")
result_handle = NCBIWWW.qblast("blastn", "nt", record.seq)
print(result_handle)from Bio import SeqIO
from Bio.Blast import NCBIWWW
record = SeqIO.read("m_cold.fasta", format="fasta")
result_handle = NCBIWWW.qblast("blastn", "nt", record.format("fasta"))
print(result_handle)셋 다 쓸 수 있다. Parse는... 그거 쓸거면 for로 불러와서 시퀀스 픽한다음 몇번쨰 시퀀스로 찾을지 골라야 하지 않나...
결과 파싱하기
from Bio import SeqIO
from Bio.Blast import NCBIWWW
record = SeqIO.read("/home/koreanraichu/CaMV.fasta", format="fasta")
result_handle = NCBIWWW.qblast("blastn", "nt", record.seq)
result_handle = open("/home/koreanraichu/CaMV_BLAST.xml")from Bio import SeqIO
from Bio.Blast import NCBIWWW
from Bio.Blast import NCBIXML
record = SeqIO.read("/home/koreanraichu/CaMV.fasta", format="fasta")
result_handle = NCBIWWW.qblast("blastn", "nt", record.seq)
blast_record = NCBIXML.read(result_handle)
print(blast_record)둘 다 해봤는데 전 아래꺼요... (위에꺼 오류남...)
save_file = open("my_blast.xml", "w") # 이거 뭐 하는 구문인데 경초 못 찾는다고 에러뜨냐
save_file.write(result_handle.read())
save_file.close()
result_handle.close()저장할거면 얘도 추가해야된다. 아까 처음 실행할 때 뭔 괴상한 문자만 땡 하고 끝났던 걸 기억하는가? Biopython에서 BLAST 돌리면 결과를 알아서 저장해주는 게 아니라 BLAST 결과에 대한 핸들을 생성한다. 그리고 그 핸들을 읽는 게 다라서 밑에 올린 저장용 코드를 따로 또 쳐 줘야 읽은 핸들을 xml파일로 저장한다.
from Bio import SeqIO
from Bio.Blast import NCBIWWW
record = SeqIO.read("/home/koreanraichu/CaMV.fasta", format="fasta")
result_handle = NCBIWWW.qblast("blastn", "nt", record.seq)
save_file = open("my_blast.xml", "w") # 이거 뭐 하는 구문인데 경로 못 찾는다고 에러뜨냐
save_file.write(result_handle.read())
save_file.close()
result_handle.close()
# 위에 네 줄까지 추가해야 결과가 저장된다.from Bio import SeqIO
from Bio.Blast import NCBIWWW
from Bio.Blast import NCBIXML
for seq_record in SeqIO.parse("/home/koreanraichu/coronavirus.gb","genbank"): # 이거 이래 불러야되나
print(seq_record.seq) # Genbank 가져올거면 이렇게 쓰세요
result_handle = NCBIWWW.qblast("blastn", "nt", seq_record.seq)
blast_record = NCBIXML.parse(result_handle)
# 결과가 하나면 read, 여러개면 parse
# 이것도 parse에 for문이 국룰인가?
save_file = open("coronavirus.xml", "w") # 이거 뭐 하는 구문인데 경로 못 찾는다고 에러뜨냐
save_file.write(result_handle.read())
save_file.close()
result_handle.close()
# 위에 네 줄까지 추가해야 결과가 저장된다근데 Genbank는 레코드 뽑는 것까지 성공했는데 BLAST 결과가 없음으로 나온다. 웹으로 한번 돌려봐야되는데 문제는 .gb파일이 안열린다는 거… (게임보이 뭐시기가 필요해서 파이참으로 바이오파이썬 불러서 열어야 한다)

아무튼 이런 식으로 저장된다. (경로 지정을 안 했더니 파이참 폴더행)
BLAST 레코드
아까 저장한 XML파일! 와! 우리꺼! 결과! 열어보자!
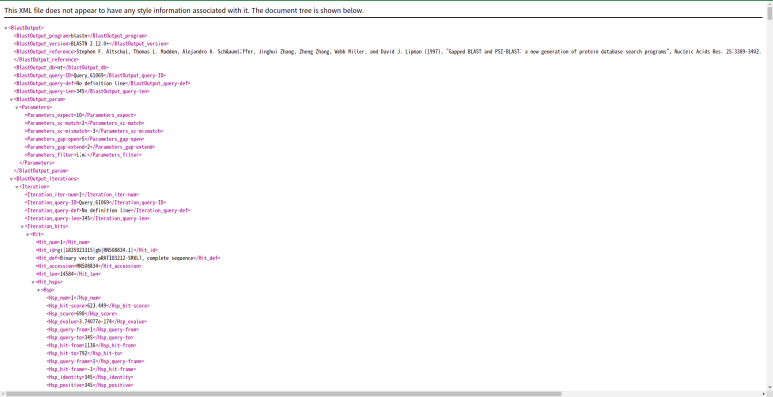
원래 저렇게 생긴 파일이다.
from Bio import SeqIO
from Bio.Blast import NCBIWWW
from Bio.Blast import NCBIXML
with open("my_blast.xml") as bf:
blast_records = NCBIXML.parse(bf)
E_VALUE_THRESH = 0.04 # 일단 이거 높으면 안좋은거다
for blast_record in blast_records:
for alignment in blast_record.alignments:
for hsp in alignment.hsps:
if hsp.expect < E_VALUE_THRESH:
print("****Alignment****")
print("sequence:", alignment.title)
print("length:", alignment.length)
print("e value:", hsp.expect)
print(hsp.query[0:75] + "...")
print(hsp.match[0:75] + "...")
print(hsp.sbjct[0:75] + "...")
# hsp 영역을 추출해 내는 스크립트.여기서 특정 영역을 추출할 수 있는데, 이 코드에서는 hsp 영역을 추출했다. 웹으로 맨날 보는 그 부분... (결과는 코드블럭에서 길다고 짤렸다...)
****Alignment****
sequence: gi|1835923315|gb|MN508834.1| Binary vector pRATIO3212-SMXL7, complete sequence
length: 14584
e value: 3.74077e-174
TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTTGCGAAGGATAGTGGGATTGTGCGTCATCCCT...
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||...
TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTTGCGAAGGATAGTGGGATTGTGCGTCATCCCT...이런 게 계~속 반복된다.
응? 벡터요? 시범조교로 벡터 쓰심? 아니 벡터가 아니라 CaMV 시퀀스 썼는데 이게 35S promoter 시퀀스였던 모양이다. CaMV는 콜리플라워 모자이크 바이러스로, 볼티모어 클래스 7(DNA-RT)에 속한다. 엔빌롭이 있고… 근데 저 바이러스가 벡터랑 뭔 상관이냐고? 유전자 과발현 벡터에는 CaMV 바이러스의 35S promoter가 들어간다.
from Bio import SeqIO
from Bio.Blast import NCBIWWW
from Bio.Blast import NCBIXML
record = SeqIO.read("/home/koreanraichu/coronavirus.gb", format="genbank")
result_handle = NCBIWWW.qblast("blastn", "nt", record.seq)
blast_records = NCBIXML.parse(result_handle)
E_VALUE_THRESH = 0.04 # 일단 이거 높으면 안좋은거다
for blast_record in blast_records:
for alignment in blast_record.alignments:
for hsp in alignment.hsps:
if hsp.expect < E_VALUE_THRESH:
print("****Alignment****")
print("sequence:", alignment.title)
print("length:", alignment.length)
print("e value:", hsp.expect)
print(hsp.query[0:75] + "...")
print(hsp.match[0:75] + "...")
print(hsp.sbjct[0:75] + "...")
# hsp 영역을 추출해 내는 스크립트.아 저장 필요없고 결과나 보여주셈! 하면 다이렉트로 돌려서 결과 보는 것도 된다. 참고로 저기 있는 E-value는 일단 클수록 안 좋다.
SearchIO
뭔 IO가 들어가요? 저거 사실 인풋/아웃풋 약어다.
아까 그 으아악 내눈을 유발했던 XML파일... 거기서 우리가 원하는 정보를 볼 수 있게 해 주는 게 이 친구 역할이다. (대충 개비스콘 아저씨 짤)
from Bio import SearchIO
blast_qresult = SearchIO.read("my_blast.xml", "blast-xml")
print(blast_qresult)Program: blastn (2.12.0+)
Query: No (345)
definition line
Target: nt
Hits: ---- ----- ----------------------------------------------------------
# # HSP ID + description
---- ----- ----------------------------------------------------------
0 1 gi|1835923315|gb|MN508834.1| Binary vector pRATIO3212-...
1 1 gi|1835923313|gb|MN508833.1| Binary vector pRATIO3212-...
2 1 gi|1835923311|gb|MN508832.1| Binary vector pRATIO1212-...
3 1 gi|1755168873|gb|MN055609.1| Plant binary expression v...
4 1 gi|1755168871|gb|MN055608.1| Plant binary expression v...
5 1 gi|1755168869|gb|MN055607.1| Plant binary expression v...
6 1 gi|1755168867|gb|MN055606.1| Plant binary expression v...
7 1 gi|1755168865|gb|MN055605.1| Plant binary expression v...
8 1 gi|1755168863|gb|MN055604.1| Plant binary expression v...
9 1 gi|1732472841|gb|MK896906.1| Plant binary expression v...
10 1 gi|1732472839|gb|MK896905.1| Plant binary expression v...
11 1 gi|1732472836|gb|MK896904.1| Plant binary expression v...
12 1 gi|1732472834|gb|MK896903.1| Plant binary expression v...
13 3 gi|1732472826|gb|MK896900.1| Plant binary expression v...
14 3 gi|1732472823|gb|MK896899.1| Plant binary expression v...
15 3 gi|1732472819|gb|MK896898.1| Plant binary expression v...
16 3 gi|1732472816|gb|MK896897.1| Plant binary expression v...
17 3 gi|1732472812|gb|MK896896.1| Plant binary expression v...
18 1 gi|1693859709|gb|MK204379.1| Binary vector pBI-Xeg, co...
19 1 gi|1624318504|gb|MK501803.1| Expression vector pHytru,...
20 1 gi|1050047859|dbj|AB830573.1| Gateway vector pB5nYGW D...
21 1 gi|1050047854|dbj|AB830572.1| Gateway vector pB5nRGW D...
22 1 gi|1050047849|dbj|AB830571.1| Gateway vector pB5nGGW D...
23 1 gi|1050047842|dbj|AB830570.1| Gateway vector pB5nCGW D...
24 1 gi|1050047835|dbj|AB830569.1| Gateway vector pB5GWnY D...
25 1 gi|1050047829|dbj|AB830568.1| Gateway vector pB5GWnR D...
26 1 gi|1050047822|dbj|AB830567.1| Gateway vector pB5GWnG D...
27 1 gi|1050047815|dbj|AB830566.1| Gateway vector pB5GWnC D...
28 1 gi|1050047807|dbj|AB830565.1| Gateway vector pB5GWcY D...
29 1 gi|1050047801|dbj|AB830564.1| Gateway vector pB5GWcR D...
~~~
47 3 gi|2087986960|gb|MW026675.1| Plant expression vector p...
48 3 gi|2033742660|gb|MW557527.1| Cloning vector pGWB-cLUC,...
49 3 gi|2033742656|gb|MW557526.1| Cloning vector pGWB-nLUC,...매우 편안한 것을 알 수 있다. 저거 판다스 데이터프레임으로 묶어서 그룹바이 마렵네
SearchIO iteration
for hit in blast_qresult:
print(hit)Query: No
definition line
Hit: gi|1835923315|gb|MN508834.1| (14584)
Binary vector pRATIO3212-SMXL7, complete sequence
HSPs: ---- -------- --------- ------ --------------- ---------------------
# E-value Bit score Span Query range Hit range
---- -------- --------- ------ --------------- ---------------------
0 3.7e-174 623.45 345 [0:345] [791:1136]저런 게 반복된다. 뭐... 리스트나 딕셔너리마냥 iteration, 인덱싱에 슬라이싱도 되고, 특정 영역(id, sequence length)만 볼 수도 있다. 물론 슬라이싱과 동시적용도 된다.
sort_key = lambda hit: hit.seq_len
sorted_qresult = blast_qresult.sort(key=sort_key, reverse=True, in_place=False)
for hit in sorted_qresult:
print("%s %i" % (hit.id, hit.seq_len))gi|1835923315|gb|MN508834.1| 14584
gi|1835923313|gb|MN508833.1| 14548
gi|1693859709|gb|MK204379.1| 13667
gi|2087986978|gb|MW026679.1| 13558
gi|2087986968|gb|MW026677.1| 13439
gi|2033742656|gb|MW557526.1| 13430
gi|2087986973|gb|MW026678.1| 13025
gi|1732472816|gb|MK896897.1| 13004
gi|2087986983|gb|MW026680.1| 12906
gi|1835923311|gb|MN508832.1| 12814
gi|1732472819|gb|MK896898.1| 12754
gi|2033742660|gb|MW557527.1| 12656
gi|1050047859|dbj|AB830573.1| 12546
gi|1050047849|dbj|AB830571.1| 12546
gi|1050047842|dbj|AB830570.1| 12546
gi|1050047835|dbj|AB830569.1| 12531
gi|1050047822|dbj|AB830567.1| 12531
gi|1050047815|dbj|AB830566.1| 12531
gi|1050047854|dbj|AB830572.1| 12519
gi|1050047829|dbj|AB830568.1| 12504
gi|2087986964|gb|MW026676.1| 12332
gi|1050047782|dbj|AB830561.1| 12266
gi|1050047801|dbj|AB830564.1| 12249
gi|1050047720|dbj|AB830552.1| 12228
gi|1050047788|dbj|AB830562.1| 12222
gi|1050047776|dbj|AB830560.1| 12222
gi|1050047770|dbj|AB830559.1| 12213
gi|1050047755|dbj|AB830557.1| 12213
gi|1050047748|dbj|AB830556.1| 12213
gi|1050047807|dbj|AB830565.1| 12207
gi|1050047794|dbj|AB830563.1| 12207
gi|1050047762|dbj|AB830558.1| 12186
gi|1050047734|dbj|AB830554.1| 11931
gi|1050047741|dbj|AB830555.1| 11889
gi|1050047727|dbj|AB830553.1| 11889
gi|2087986960|gb|MW026675.1| 11799
gi|1732472826|gb|MK896900.1| 11186
gi|1732472823|gb|MK896899.1| 11185
gi|1732472812|gb|MK896896.1| 10473
gi|1732472836|gb|MK896904.1| 6122
gi|1755168865|gb|MN055605.1| 5507
gi|1732472841|gb|MK896906.1| 5507
gi|1755168863|gb|MN055604.1| 5506
gi|1732472839|gb|MK896905.1| 5506
gi|1755168869|gb|MN055607.1| 5498
gi|1755168867|gb|MN055606.1| 5497
gi|1755168873|gb|MN055609.1| 5465
gi|1755168871|gb|MN055608.1| 5464
gi|1624318504|gb|MK501803.1| 4844
gi|1732472834|gb|MK896903.1| 4794판다스마냥 특정 요소를 기준으로 정렬할 수도 있다. (시퀀스 길이 기준으로 정렬한 것)
filter_func = lambda hit: len(hit.hsps) > 1
filtered_qresult = blast_qresult.hit_filter(filter_func)
print(filtered_qresult)Query: No (345)
definition line
Target: nt
Hits: ---- ----- ----------------------------------------------------------
# # HSP ID + description
---- ----- ----------------------------------------------------------
0 3 gi|1732472826|gb|MK896900.1| Plant binary expression v...
1 3 gi|1732472823|gb|MK896899.1| Plant binary expression v...
2 3 gi|1732472819|gb|MK896898.1| Plant binary expression v...
3 3 gi|1732472816|gb|MK896897.1| Plant binary expression v...
4 3 gi|1732472812|gb|MK896896.1| Plant binary expression v...
5 3 gi|2087986983|gb|MW026680.1| Plant expression vector p...
6 3 gi|2087986978|gb|MW026679.1| Plant expression vector p...
7 3 gi|2087986973|gb|MW026678.1| Plant expression vector p...
8 3 gi|2087986968|gb|MW026677.1| Plant expression vector p...
9 3 gi|2087986964|gb|MW026676.1| Plant expression vector p...
10 3 gi|2087986960|gb|MW026675.1| Plant expression vector p...
11 3 gi|2033742660|gb|MW557527.1| Cloning vector pGWB-cLUC,...
12 3 gi|2033742656|gb|MW557526.1| Cloning vector pGWB-nLUC,...
Process finished with exit code 0함수 걸어서 필터링도 된다는데 람다가 국룰인건가...
Hit, HSP, Fragment
무슨 족보 그리는것도 아니고 하위의 하위 뭔데...
Hit
아까 hit.id 이런 식으로 잠깐 지나쳤던 것. 어쨌거나 상위 개념인 듯 하다.
from Bio import SearchIO
blast_qresult = SearchIO.read("my_blast.xml", "blast-xml")
blast_hit = blast_qresult[0]
print(blast_hit)Query: No
definition line
Hit: gi|1835923315|gb|MN508834.1| (14584)
Binary vector pRATIO3212-SMXL7, complete sequence
HSPs: ---- -------- --------- ------ --------------- ---------------------
# E-value Bit score Span Query range Hit range
---- -------- --------- ------ --------------- ---------------------
0 3.7e-174 623.45 345 [0:345] [791:1136]색인할 수 있다고 한다. 참고로 저 [0]을 잘 기억해두자.
HSP
힛샥 프로테인...아니고 High scoring pair의 준말이다.
from Bio import SearchIO
blast_qresult = SearchIO.read("my_blast.xml", "blast-xml")
blast_hsp = blast_qresult[0][0]
print(blast_hsp)Query: No definition line
Hit: gi|1835923315|gb|MN508834.1| Binary vector pRATIO3212-SMXL7, com...
Query range: [0:345] (1)
Hit range: [791:1136] (-1)
Quick stats: evalue 3.7e-174; bitscore 623.45
Fragments: 1 (345 columns)
Query - TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTTGCGAAGGATAGTGGG~~~TCAAT
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~|||||
Hit - TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTTGCGAAGGATAGTGGG~~~TCAAT
Process finished with exit code 0얘는 그래서 힛이랑 달리 색인할 때 리스트 두 개가 들어간다. 각각 맨 첫번째 힛에서 맨 첫번째 HSP를 뜻한다. 색인할 때 하나 빼먹으면 힛이 출력된다.
# 얘네는 hsp 위치까지 지정해줘야 출력한다
print(blast_hsp.query_range) # 첫번째 hit의 첫번째 hsp의 query range
print(blast_hsp.evalue) # 첫번째 hit의 첫번째 hsp의 evalue
print(blast_hsp.hit_start) # 첫번째 hit의 첫번째 hsp의 hit start coordinates
print(blast_hsp.query_span) # 첫번째 hit의 첫번째 hsp의 how many residues in the query sequence(residue 몇개냐)
print(blast_hsp.aln_span) # 첫번째 hit의 첫번째 hsp의 alignment 길이
print(blast_hsp.gap_num) # 첫번째 hit의 첫번째 hsp의 gap no.
print(blast_hsp.ident_num) # 첫번째 hit의 첫번째 hsp의 identical residue no.
print(blast_hsp.aln) # 배열 뽑아줘!(0, 345)
3.74077e-174
791
345
345
0
345
# 밑에는 배열이다
Alignment with 2 rows and 345 columns
TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTT...AAT No
TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTT...AAT gi|1835923315|gb|MN508834.1|참고로 힛 상태에서 저거 쓰면 오류난다.
HSPFragment
가장 하위 개념. 얘는 색인할 때 []가 세 개 들어간다.
blast_frag = blast_qresult[0][0][0] # 첫번째 hit의 첫번째 hsp의 첫번째 fragment
print(blast_frag)Query: No definition line
Hit: gi|1835923315|gb|MN508834.1| Binary vector pRATIO3212-SMXL7, com...
Query range: [0:345] (1)
Hit range: [791:1136] (-1)
Fragments: 1 (345 columns)
Query - TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTTGCGAAGGATAGTGGG~~~TCAAT
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||~~~|||||
Hit - TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTTGCGAAGGATAGTGGG~~~TCAAT
Process finished with exit code 0...HSP랑 차이 없는데?
blast_frag = blast_qresult[0][0][0] # 첫번째 hit의 첫번째 hsp의 첫번째 fragment
print(blast_frag.hit) # hit sequence(sequence object)
print(blast_frag.query_start) # 시작 좌표
print(blast_frag.hit_strand) # hi sequence strand 수ID: gi|1835923315|gb|MN508834.1|
Name: aligned hit sequence
Description: Binary vector pRATIO3212-SMXL7, complete sequence
Number of features: 0
/molecule_type=DNA
Seq('TCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTTGCGAAGGATA...AAT')
0
-1아니 strand 수에 -1은 뭐여...
'Coding > Python' 카테고리의 다른 글
| Biopython으로 Swiss-prot과 ExPASy 데이터베이스 탐방하기 (0) | 2022.08.21 |
|---|---|
| Biopython으로 Entrez database 탐방하기 (0) | 2022.08.21 |
| 번외편-코딩테스트 풀이 (0) | 2022.08.20 |
| Biopython으로 MSA 해보기 (0) | 2022.08.20 |
| Biopython SeqIO 써보기 (0) | 2022.08.20 |



